Cas12a-based rapid method for early detection of X-disease phytoplasma
Author: Dr. Youfu “Frank” Zhao
Published: 2025
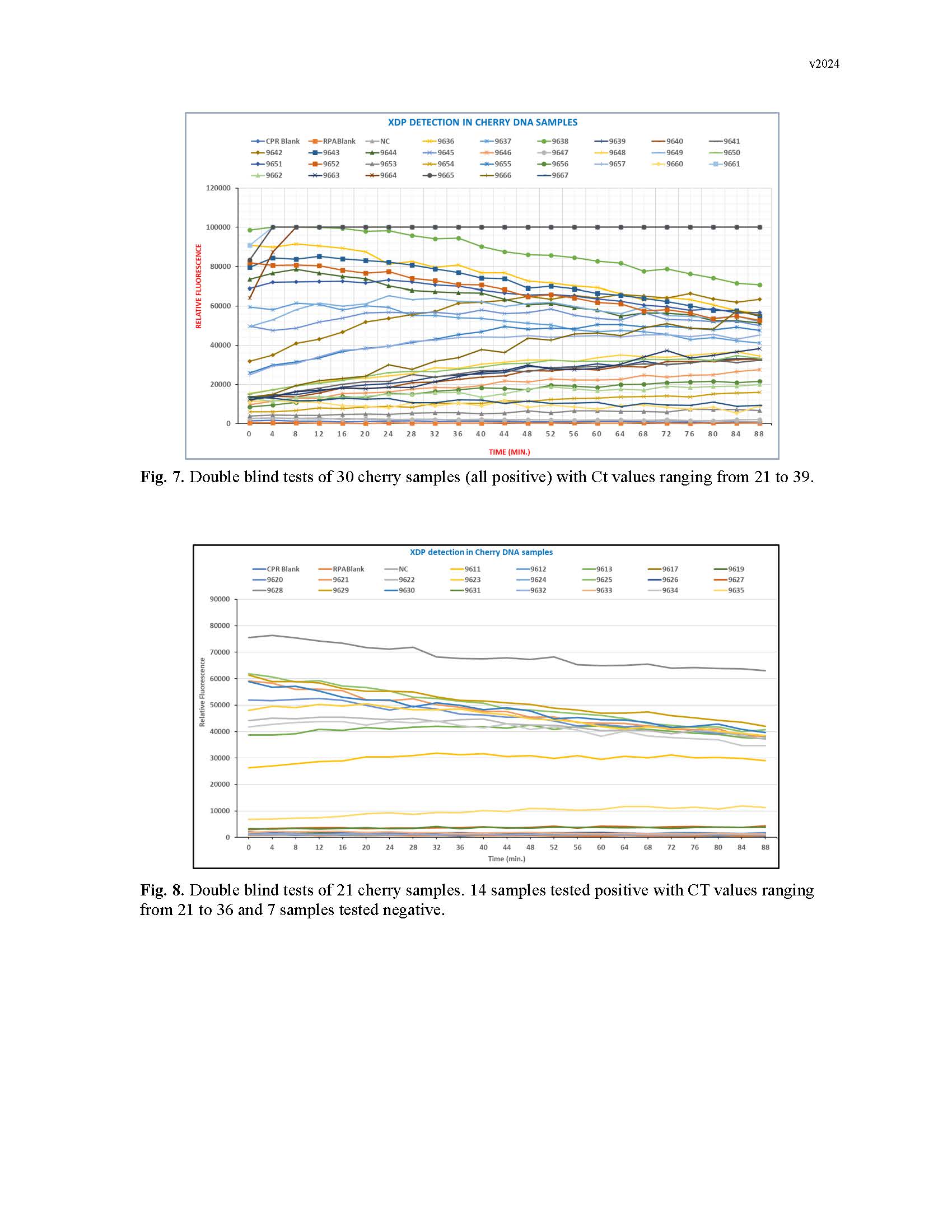
Summary: The cherry X-disease is at epidemic levels in the Pacific Northwest and caused significant economic losses in the past several years. To effectively manage the X-disease phytoplasma (XDP), it is critical that infected trees be identified as rapidly as possible, especially before symptoms appear. Recently, the clustered regularly interspersed short palindromic repeats/CRISPA-associated (CRISPR-Cas) system has been used as an extremely sensitive and rapid diagnostic tool for pathogen detection besides its common applications in genome editing. In this study, we have developed the recombinase polymerase amplification (RPA)/Cas12a diagnostic assay and a quick DNA extraction method to facilitate the rapid and super-sensitive detection of XDP. Based on the DNA sequence alignment and analysis of the SecY genes from the phytoplasma 16SrIII subgroups, XDP-specific RPA primers and crRNAs have been identified and validated for its high efficiency of RPA and for the XDP Cas12a detection assay. Thus, a two-pot RPA/Cas12a assay has been successfully developed for highly sensitive and specific detection of different XDP strains and demonstrated to be capable of specific and rapid detection of XDP-infected samples from the field. Double blind assay results from Cas12a-based tests for more than 100 samples of sweet cherries, weeds and insects correlated well with the current industry standard qPCR assay results. In addition, we also developed a 5-min genomic DNA extraction method for the RPA/Cas12a assay and established one-pot RPA/Cas12a assay based on the cap/spin approach, which will facilitate the field-deployable detection of XDP in the near future. In summary, the highly sensitive, specific, and rapid RPA/Cas12a assay for XDP detection not only provides the industry a much-needed platform for fast, supersensitive, and accurate diagnosis, but also saves time and money for both growers and nurseries.
Keywords: